Run GTEx v8 data on Terra
Chun-Jie Liu · 2022-05-09
Background #
GTEx v8 protected data require an approved dbGaP application. GTEx v8 protected data are now hosted in the AnVIL repository, with access granted to users who have approved dbGaP applications.
To access GTEx v8 in AnVIL:
- Approved dbGaP GTEx.
- Create an account on AnVIL.
- Link eRA Commons ID to Terra account.
- The data for GTEx v8 is found in workspace located here.
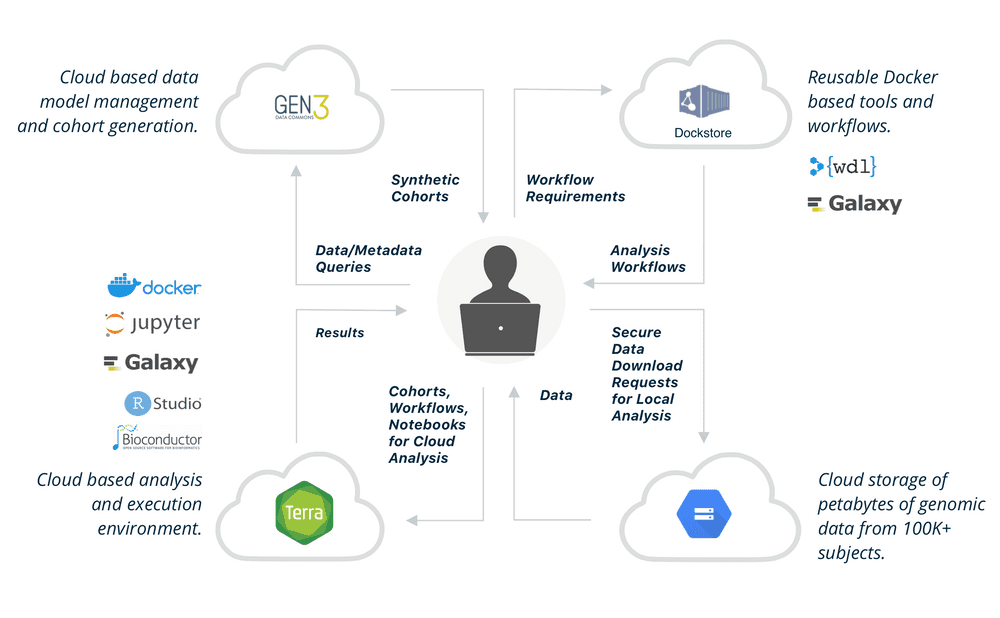
AnVIL is NHGRI’s Genomic Data Science Analysis, Visualization, and Informatics Lab-Space. AnVIL inverts the traditional model, providing a cloud environment for the analysis of large genomic and related datasets. By providing a unified environment for data management and compute, AnVIL eliminates the need for data movement, allows for active threat detect and monitoring, and provide elastic, shared computing resources that can be acquired by researcers.
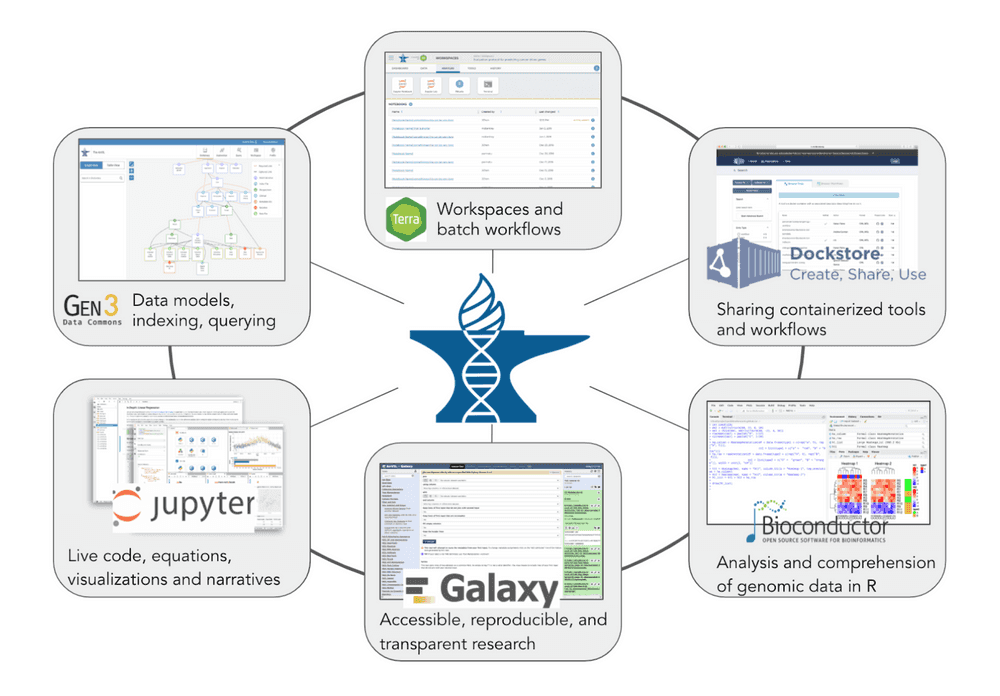
AnVIL uses Terra as its analysis platform, Gen3 for data search and artificial cohort creation, and Dockstore as a repository for Docker-based genomic analysis tools and workflows.
The platform is built on a set of established components. The Terra platform provides environment with secure data and analysis sharing capabilities, it is powered by Google Cloud Platform. Dockstore provides standards based sharing of containerized tools and workflows. The Gen3 data commons framework provides data and metadata ingest, querying, and organization.
Setup running environment on Terra #
After registering on the Terra and link to NIH eRA Commons ID, you can access the GTEx data in AnVIL. Then creating a billing account on GCP and link to Terra billing account follow the instractions. GCP provides $300 credits trial for new users, set up a billing budget to your GCP billing.
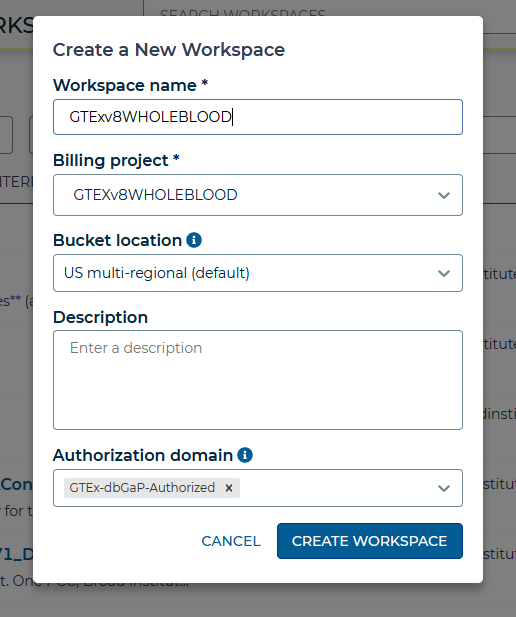
Next, creating a workspace on Terra to select billing project and authorization domain. Then add your data and running pipeline to the workspace.
Test sQTL analysis workflow on GTEX v8 WHOLE BLOOD sub datasets. #
Prepare data #
- Download data table from AvVIL_GTEXv8_hg38 data https://anvil.terra.bio/#workspaces/anvil-datastorage/AnVIL_GTEx_V8_hg38/data. And filter the whole blood samples. Then upload whole blood samples and participant to the data table.
- Add reference data used by GTEx v8 to the Workspace Data.
- GTEX v8 whole blood dataset contains 755 samples.
- The samples was compressed as bam files.
- Call splice event directly from these bam files.
Prepare workflow #
- Modify rMATS-Turbo WDL workflow for One Group analysis.
- Push the WDL workflow to the dockstore.
- Link this WDL workflow to Terra.
- Update Inputs with required parameters and click start run.
Test with small sample size #
- Select first two samples to test the worflow.
readLength of the bam files, the average readLength of GTEx v8 Whole Blood tissue RNA-seq data is 152, but some samples' readLength is 136, thus variable_readLength is recommended to set as TRUE.
WDL workflow takes the list of samples as a job, but the Terra sample data table regard each sample as a job. We need to reorganize the data table to sample_set and then use the sample bam list as input.
Run on all 755 GTEx v8 whole blood tissue samples. #
- Upload sample.tsv, participant.tsv and sample_set.tsv
- Start to run.